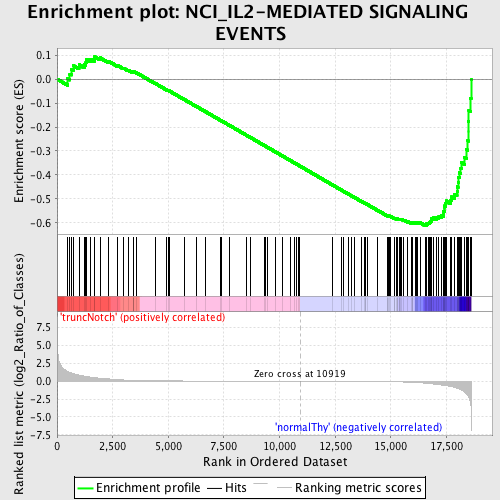
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | NCI_IL2-MEDIATED SIGNALING EVENTS |
| Enrichment Score (ES) | -0.6131057 |
| Normalized Enrichment Score (NES) | -1.5425882 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.33624274 |
| FWER p-Value | 0.939 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RPS6 | 2060168 | 478 | 1.354 | 0.0014 | No | ||
| 2 | MAPKAPK2 | 2850500 | 572 | 1.240 | 0.0213 | No | ||
| 3 | IL2RA | 6620450 | 647 | 1.126 | 0.0399 | No | ||
| 4 | STAT5A | 2680458 | 724 | 1.049 | 0.0568 | No | ||
| 5 | JAK3 | 70347 3290008 | 985 | 0.859 | 0.0600 | No | ||
| 6 | SOCS3 | 5550563 | 1234 | 0.696 | 0.0606 | No | ||
| 7 | ARRB2 | 2060441 | 1293 | 0.658 | 0.0707 | No | ||
| 8 | E2F1 | 5360093 | 1330 | 0.638 | 0.0816 | No | ||
| 9 | HRAS | 1980551 | 1508 | 0.558 | 0.0832 | No | ||
| 10 | MAPK3 | 580161 4780035 | 1667 | 0.499 | 0.0847 | No | ||
| 11 | PRKCZ | 3780279 | 1695 | 0.488 | 0.0930 | No | ||
| 12 | NFKB1 | 5420358 | 1928 | 0.422 | 0.0890 | No | ||
| 13 | GRB2 | 6650398 | 2319 | 0.313 | 0.0742 | No | ||
| 14 | PRF1 | 6660309 | 2711 | 0.213 | 0.0573 | No | ||
| 15 | IL2RB | 4730072 | 2982 | 0.168 | 0.0461 | No | ||
| 16 | BCL3 | 3990440 | 3203 | 0.139 | 0.0370 | No | ||
| 17 | PPP2R5D | 4010156 380408 5550112 5670162 | 3414 | 0.116 | 0.0280 | No | ||
| 18 | MAP2K2 | 4590601 | 3424 | 0.115 | 0.0298 | No | ||
| 19 | SMPD1 | 1410309 5550717 | 3437 | 0.114 | 0.0315 | No | ||
| 20 | PRKCA | 6400551 | 3576 | 0.103 | 0.0261 | No | ||
| 21 | NRAS | 6900577 | 4430 | 0.057 | -0.0189 | No | ||
| 22 | RPS6KB1 | 1450427 5080110 6200563 | 4932 | 0.043 | -0.0451 | No | ||
| 23 | HSP90AA1 | 4560041 5220133 2120722 | 4987 | 0.042 | -0.0472 | No | ||
| 24 | IL4 | 6020537 | 5032 | 0.041 | -0.0487 | No | ||
| 25 | FRAP1 | 2850037 6660010 | 5731 | 0.028 | -0.0859 | No | ||
| 26 | CALM1 | 380128 | 6258 | 0.021 | -0.1139 | No | ||
| 27 | CDK6 | 4920253 | 6691 | 0.017 | -0.1369 | No | ||
| 28 | PIK3R1 | 4730671 | 7321 | 0.013 | -0.1706 | No | ||
| 29 | FOS | 1850315 | 7390 | 0.012 | -0.1741 | No | ||
| 30 | SOS1 | 7050338 | 7726 | 0.010 | -0.1920 | No | ||
| 31 | PTPN11 | 2230100 2470180 6100528 | 8524 | 0.007 | -0.2349 | No | ||
| 32 | REL | 360707 | 8673 | 0.007 | -0.2428 | No | ||
| 33 | AKT1 | 5290746 | 9329 | 0.005 | -0.2781 | No | ||
| 34 | RAN | 2260446 4590647 | 9367 | 0.004 | -0.2800 | No | ||
| 35 | XPO1 | 540707 | 9449 | 0.004 | -0.2843 | No | ||
| 36 | IL2 | 1770725 | 9837 | 0.003 | -0.3051 | No | ||
| 37 | GAB2 | 1410280 2340520 4280040 | 10137 | 0.002 | -0.3212 | No | ||
| 38 | PRKCE | 5700053 | 10501 | 0.001 | -0.3408 | No | ||
| 39 | INSL3 | 4150092 | 10652 | 0.001 | -0.3489 | No | ||
| 40 | MYC | 380541 4670170 | 10764 | 0.000 | -0.3549 | No | ||
| 41 | SGMS1 | 4780195 | 10842 | 0.000 | -0.3591 | No | ||
| 42 | ATM | 3610110 4050524 | 10891 | 0.000 | -0.3616 | No | ||
| 43 | TNF | 6650603 | 12391 | -0.005 | -0.4426 | No | ||
| 44 | TERT | 6660075 | 12800 | -0.007 | -0.4645 | No | ||
| 45 | IFNG | 5670592 | 12891 | -0.007 | -0.4692 | No | ||
| 46 | FBXW11 | 6450632 | 13100 | -0.009 | -0.4802 | No | ||
| 47 | SRC | 580132 | 13235 | -0.010 | -0.4873 | No | ||
| 48 | IRS1 | 1190204 | 13369 | -0.011 | -0.4943 | No | ||
| 49 | SYK | 6940133 | 13689 | -0.014 | -0.5112 | No | ||
| 50 | FOXO3 | 2510484 4480451 | 13812 | -0.016 | -0.5175 | No | ||
| 51 | JUN | 840170 | 13846 | -0.016 | -0.5189 | No | ||
| 52 | PIK3CA | 6220129 | 13861 | -0.016 | -0.5194 | No | ||
| 53 | STAT5B | 6200026 | 13947 | -0.018 | -0.5236 | No | ||
| 54 | ERC1 | 1580170 2630717 6110762 6840280 | 14397 | -0.028 | -0.5473 | No | ||
| 55 | CCNA2 | 5290075 | 14835 | -0.048 | -0.5699 | No | ||
| 56 | NOD2 | 2510050 | 14872 | -0.050 | -0.5709 | No | ||
| 57 | STAM | 3850008 4760441 | 14899 | -0.052 | -0.5712 | No | ||
| 58 | MALT1 | 4670292 | 14919 | -0.053 | -0.5712 | No | ||
| 59 | STAM2 | 730731 2120377 | 14969 | -0.055 | -0.5727 | No | ||
| 60 | MAPK11 | 130452 3120440 3610465 | 15150 | -0.068 | -0.5811 | No | ||
| 61 | FASLG | 2810044 | 15163 | -0.069 | -0.5804 | No | ||
| 62 | RELB | 1400048 | 15248 | -0.075 | -0.5834 | No | ||
| 63 | LTA | 1740088 | 15289 | -0.079 | -0.5840 | No | ||
| 64 | SHC1 | 2900731 3170504 6520537 | 15309 | -0.081 | -0.5834 | No | ||
| 65 | IKBKG | 3450092 3840377 6590592 | 15397 | -0.091 | -0.5863 | No | ||
| 66 | JAK1 | 5910746 | 15408 | -0.091 | -0.5850 | No | ||
| 67 | BIRC2 | 2940435 4730020 | 15427 | -0.093 | -0.5841 | No | ||
| 68 | MAP2K1 | 840739 | 15475 | -0.098 | -0.5847 | No | ||
| 69 | NFKB2 | 2320670 | 15588 | -0.110 | -0.5885 | No | ||
| 70 | FYN | 2100468 4760520 4850687 | 15761 | -0.130 | -0.5952 | No | ||
| 71 | UBE2D3 | 3190452 | 15912 | -0.149 | -0.6003 | No | ||
| 72 | MAPK1 | 3190193 6200253 | 15968 | -0.160 | -0.6001 | No | ||
| 73 | BCL2 | 730132 1570736 2470138 3800044 4810037 5690068 5860504 6650164 | 15995 | -0.163 | -0.5982 | No | ||
| 74 | RAC1 | 4810687 | 16093 | -0.176 | -0.5999 | No | ||
| 75 | RIPK2 | 5050072 6290632 | 16153 | -0.187 | -0.5993 | No | ||
| 76 | DOK2 | 5340273 | 16176 | -0.191 | -0.5966 | No | ||
| 77 | FOXP3 | 940707 1850692 | 16316 | -0.219 | -0.5998 | No | ||
| 78 | MAPK8 | 2640195 | 16564 | -0.281 | -0.6075 | Yes | ||
| 79 | IKBKB | 6840072 | 16617 | -0.298 | -0.6043 | Yes | ||
| 80 | LCK | 3360142 | 16671 | -0.311 | -0.6009 | Yes | ||
| 81 | CALM2 | 6620463 | 16731 | -0.330 | -0.5975 | Yes | ||
| 82 | SP1 | 6590017 | 16789 | -0.347 | -0.5936 | Yes | ||
| 83 | CDK2 | 130484 2260301 4010088 5050110 | 16829 | -0.357 | -0.5885 | Yes | ||
| 84 | RELA | 3830075 | 16834 | -0.358 | -0.5815 | Yes | ||
| 85 | RAF1 | 1770600 | 16929 | -0.388 | -0.5788 | Yes | ||
| 86 | CYLD | 6590079 | 17073 | -0.438 | -0.5777 | Yes | ||
| 87 | KRAS | 2060170 | 17160 | -0.476 | -0.5728 | Yes | ||
| 88 | MYB | 1660494 5860451 6130706 | 17283 | -0.524 | -0.5689 | Yes | ||
| 89 | CALM3 | 3390288 | 17358 | -0.561 | -0.5616 | Yes | ||
| 90 | CHUK | 7050736 | 17374 | -0.567 | -0.5510 | Yes | ||
| 91 | TRAF6 | 4810292 6200132 | 17407 | -0.581 | -0.5411 | Yes | ||
| 92 | MAPK14 | 5290731 | 17410 | -0.583 | -0.5295 | Yes | ||
| 93 | NFKBIA | 1570152 | 17455 | -0.610 | -0.5196 | Yes | ||
| 94 | SOCS2 | 4760692 | 17482 | -0.624 | -0.5085 | Yes | ||
| 95 | RASA1 | 1240315 | 17696 | -0.741 | -0.5051 | Yes | ||
| 96 | BCL10 | 2360397 | 17734 | -0.764 | -0.4918 | Yes | ||
| 97 | MAP3K14 | 5890435 | 17867 | -0.866 | -0.4815 | Yes | ||
| 98 | SOCS1 | 730139 | 17976 | -0.958 | -0.4681 | Yes | ||
| 99 | IL2RG | 4120273 | 17992 | -0.973 | -0.4494 | Yes | ||
| 100 | STAT1 | 6510204 6590553 | 18024 | -1.010 | -0.4308 | Yes | ||
| 101 | STAT3 | 460040 3710341 | 18039 | -1.029 | -0.4109 | Yes | ||
| 102 | PTK2B | 4730411 | 18075 | -1.069 | -0.3913 | Yes | ||
| 103 | BCL2L1 | 1580452 4200152 5420484 | 18144 | -1.180 | -0.3713 | Yes | ||
| 104 | ELF1 | 4780450 | 18178 | -1.217 | -0.3486 | Yes | ||
| 105 | UGCG | 3420053 | 18307 | -1.492 | -0.3255 | Yes | ||
| 106 | MAPK9 | 2060273 3780209 4070397 | 18410 | -1.795 | -0.2950 | Yes | ||
| 107 | TNFAIP3 | 2900142 | 18460 | -1.987 | -0.2578 | Yes | ||
| 108 | CISH | 840315 | 18478 | -2.069 | -0.2171 | Yes | ||
| 109 | CCND3 | 380528 730500 | 18479 | -2.071 | -0.1755 | Yes | ||
| 110 | RHOA | 580142 5900131 5340450 | 18507 | -2.308 | -0.1306 | Yes | ||
| 111 | CCND2 | 5340167 | 18568 | -2.711 | -0.0794 | Yes | ||
| 112 | TNFRSF1A | 1090390 6520735 | 18609 | -4.080 | 0.0004 | Yes |

